Introduction to Model Validation
Model validation answers the crucial question: "Will this model work in the real world?" Diagnostic plots tell you if assumptions are met, but only testing on new data reveals true predictive performance.
What You'll Learn
- Implement leave-one-out and k-fold cross-validation
- Assess prediction accuracy with multiple metrics
- Use bootstrap methods for uncertainty quantification
- Understand the bias-variance trade-off in model complexity
- Distinguish between confidence and prediction intervals
- Apply learning curves to assess data sufficiency
- Make informed decisions about model deployment
- Completed modeling, comparison, and diagnostic tutorials
- Understanding of overfitting and generalization
- Same packages:
tidyverse,glmmTMB,broom.mixed - Basic knowledge of sampling and resampling concepts
Cross-Validation: The Gold Standard
Cross-validation tests model performance by repeatedly fitting on subsets of data and predicting on held-out observations. This simulates real-world deployment where you must predict new cases.
# Implement Leave-One-Out Cross-Validation
library(tidyverse)
library(glmmTMB)
library(broom.mixed)
data(mtcars)
# Define our competing models from previous lessons
loo_cv <- function(model_formula, data) {
n <- nrow(data)
predictions <- numeric(n)
for(i in 1:n) {
# Fit model without observation i
train_data <- data[-i, ]
test_data <- data[i, ]
# Fit model and predict held-out observation
temp_model <- glmmTMB(model_formula, data = train_data)
predictions[i] <- predict(temp_model, newdata = test_data)
}
return(predictions)
}
# Test all our competing models
cv_formulas <- list(
simple = mpg ~ wt,
additive = mpg ~ wt + hp,
interaction = mpg ~ wt * hp,
complex = mpg ~ wt * hp + cyl
)
# Fit all models to the full dataset for AIC calculation
# Note: These fitted models are needed for AIC comparison and bootstrap methods
models <- list(
simple = glmmTMB(cv_formulas$simple, data = mtcars),
additive = glmmTMB(cv_formulas$additive, data = mtcars),
interaction = glmmTMB(cv_formulas$interaction, data = mtcars),
complex = glmmTMB(cv_formulas$complex, data = mtcars)
)
# Perform cross-validation for each model
cv_results <- map_dfr(names(cv_formulas), function(model_name) {
predictions <- loo_cv(cv_formulas[[model_name]], mtcars)
data.frame(
model = model_name,
observed = mtcars$mpg,
predicted = predictions,
residual = mtcars$mpg - predictions,
car = rownames(mtcars)
)
})
# Calculate performance metrics
cv_metrics <- cv_results %>%
group_by(model) %>%
summarise(
rmse = sqrt(mean(residual^2)),
mae = mean(abs(residual)),
r_squared = cor(observed, predicted)^2,
.groups = 'drop'
)
print(cv_metrics)| Model | RMSE | MAE | R-squared |
|---|---|---|---|
| Simple (wt only) | 3.20 | 2.52 | 0.710 |
| Additive (wt + hp) | 2.78 | 2.12 | 0.783 |
| Interaction (wt × hp) | 2.34 | 2.03 | 0.845 |
| Complex (wt × hp + cyl) | 2.38 | 2.06 | 0.840 |
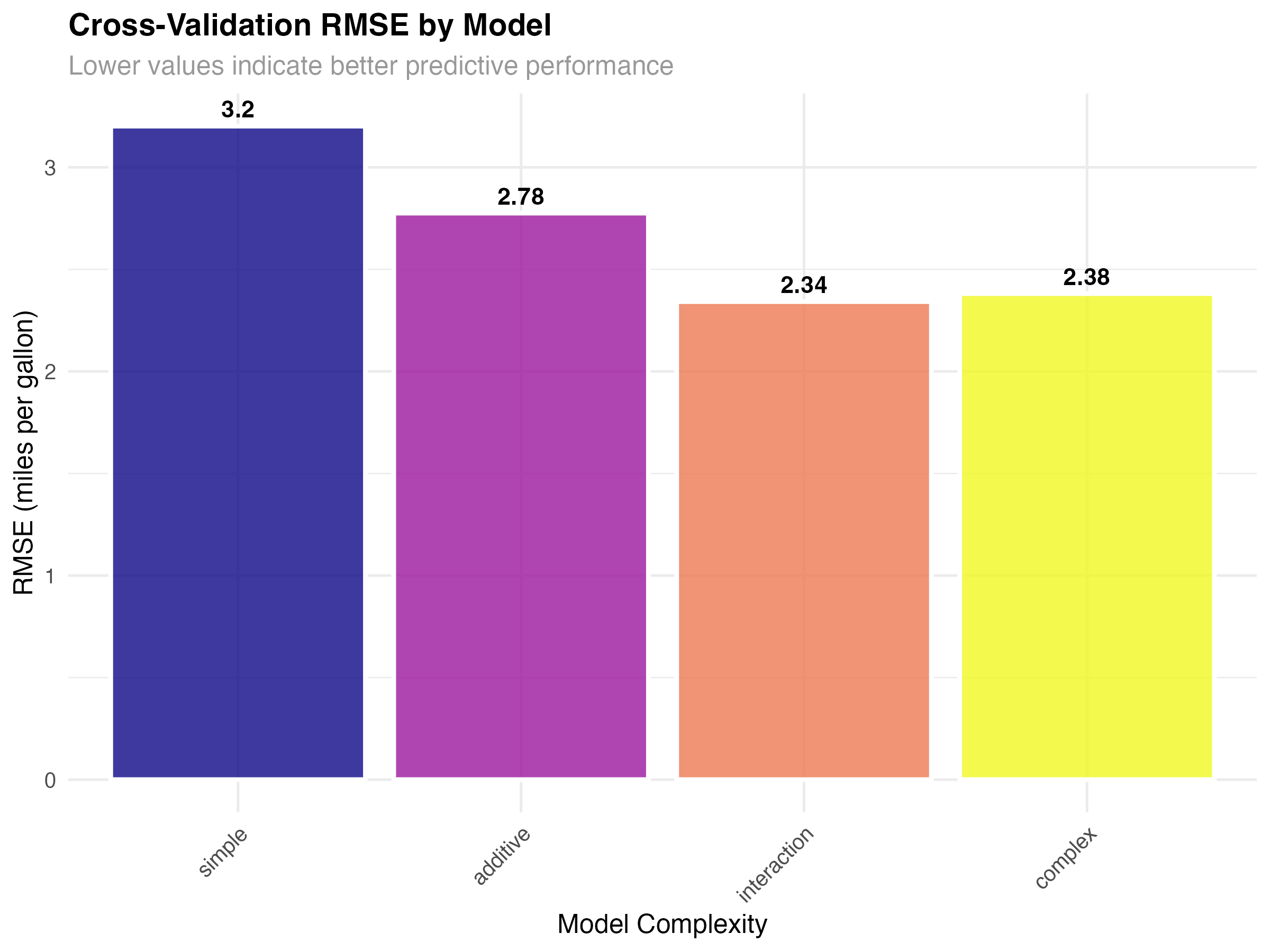
Understanding the Results
- Simple model (RMSE: 3.20): Underfit - misses important relationships
- Additive model (RMSE: 2.78): Better but still missing interaction effects
- Interaction model (RMSE: 2.34): Best balance of complexity and generalization
- Complex model (RMSE: 2.38): Slight overfitting - worse than simpler interaction model
# Visualize prediction accuracy for best model
best_model_cv <- cv_results %>% filter(model == "interaction")
ggplot(best_model_cv, aes(x = observed, y = predicted)) +
geom_point(size = 3, alpha = 0.7, color = "#2563eb") +
geom_smooth(method = "lm", se = TRUE, color = "#dc2626", linewidth = 1.2) +
geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "gray50") +
geom_text(aes(label = car), size = 2.5, nudge_y = 0.8, alpha = 0.7) +
labs(
title = "Cross-Validation: Predicted vs Observed MPG",
subtitle = "Interaction model - Points closer to diagonal indicate better predictions",
x = "Observed MPG",
y = "Predicted MPG"
) +
theme_minimal() +
coord_equal()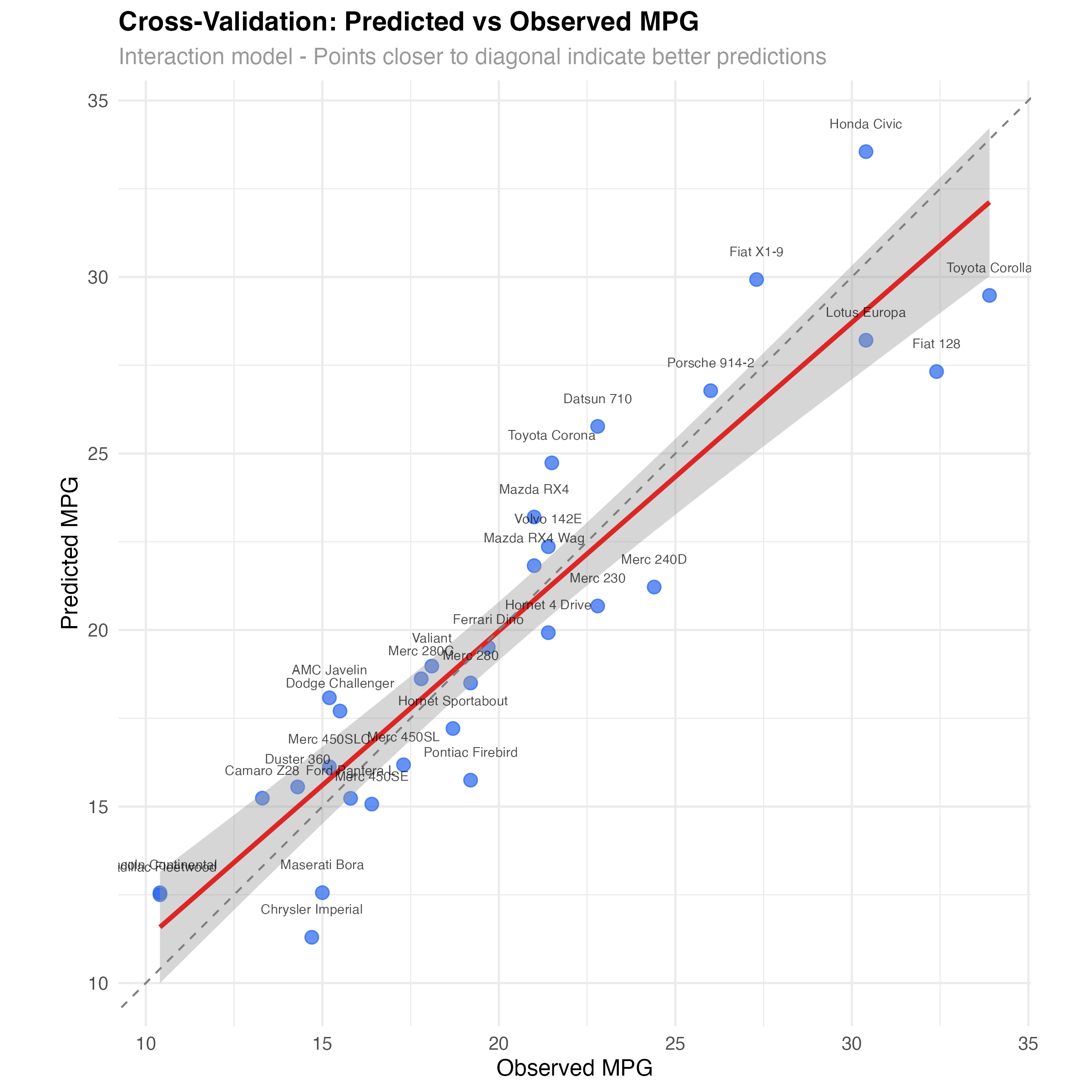
Prediction Error Analysis
Beyond overall metrics, examining the distribution of prediction errors reveals which types of observations are difficult to predict and whether errors are systematic.
# Analyze prediction error patterns
ggplot(cv_results, aes(x = model, y = abs(residual), fill = model)) +
geom_boxplot(alpha = 0.7, outlier.alpha = 0.6) +
geom_jitter(width = 0.2, alpha = 0.5, size = 1.5) +
scale_fill_viridis_d(option = "plasma") +
labs(
title = "Cross-Validation Residual Magnitude by Model",
subtitle = "Distribution of absolute prediction errors",
x = "Model Complexity",
y = "Absolute Residual (MPG)"
) +
theme_minimal() +
theme(legend.position = "none")
Error Pattern Analysis
- Simple model: Largest errors and widest spread - consistent underfitting
- Additive model: Reduced error spread but still some large residuals
- Interaction model: Smallest median error and tightest distribution
- Complex model: Slightly worse than interaction - overfitting penalty
Identifying Difficult-to-Predict Cases
# Find cars with consistently large prediction errors
error_analysis <- cv_results %>%
group_by(car) %>%
summarise(
mean_abs_error = mean(abs(residual)),
worst_model = model[which.max(abs(residual))],
best_model = model[which.min(abs(residual))],
.groups = 'drop'
) %>%
arrange(desc(mean_abs_error))
# Top 5 hardest to predict cars
print(head(error_analysis, 5))
# Cars that benefit most from complex models
complexity_benefit <- cv_results %>%
select(car, model, residual) %>%
pivot_wider(names_from = model, values_from = residual) %>%
mutate(
simple_to_interaction = abs(simple) - abs(interaction),
interaction_benefit = simple_to_interaction > 1 # More than 1 MPG improvement
)
print(filter(complexity_benefit, interaction_benefit))Bootstrap Uncertainty Quantification
Bootstrap resampling provides another perspective on model uncertainty by repeatedly fitting models to random samples of your data, revealing how stable your predictions are.
# Bootstrap confidence intervals for predictions
bootstrap_predictions <- function(model, newdata, n_boot = 100) {
boot_preds <- replicate(n_boot, {
# Bootstrap sample
boot_indices <- sample(nrow(mtcars), replace = TRUE)
boot_data <- mtcars[boot_indices, ]
# Fit model to bootstrap sample
boot_model <- glmmTMB(mpg ~ wt * hp, data = boot_data)
# Predict on new data
predict(boot_model, newdata = newdata)
})
# Calculate confidence intervals
data.frame(
predicted = rowMeans(boot_preds),
lower = apply(boot_preds, 1, quantile, 0.025),
upper = apply(boot_preds, 1, quantile, 0.975)
)
}
# Create prediction grid
pred_grid <- expand_grid(
wt = seq(2, 5, by = 0.5),
hp = c(100, 200, 300)
)
# Get bootstrap predictions
boot_results <- bootstrap_predictions(models$interaction, pred_grid)
pred_grid <- bind_cols(pred_grid, boot_results)
# Visualize bootstrap uncertainty
ggplot(pred_grid, aes(x = wt, y = predicted, color = factor(hp))) +
geom_ribbon(aes(ymin = lower, ymax = upper, fill = factor(hp)),
alpha = 0.2, color = NA) +
geom_line(linewidth = 1.2) +
geom_point(data = mtcars, aes(x = wt, y = mpg),
color = "black", size = 2, alpha = 0.7, inherit.aes = FALSE) +
scale_color_viridis_d(name = "Horsepower", option = "plasma") +
scale_fill_viridis_d(name = "Horsepower", option = "plasma") +
labs(
title = "Bootstrap Confidence Intervals for Predictions",
subtitle = "Model uncertainty quantified through resampling",
x = "Weight (1000 lbs)",
y = "Predicted MPG"
) +
theme_minimal() +
theme(legend.position = "bottom")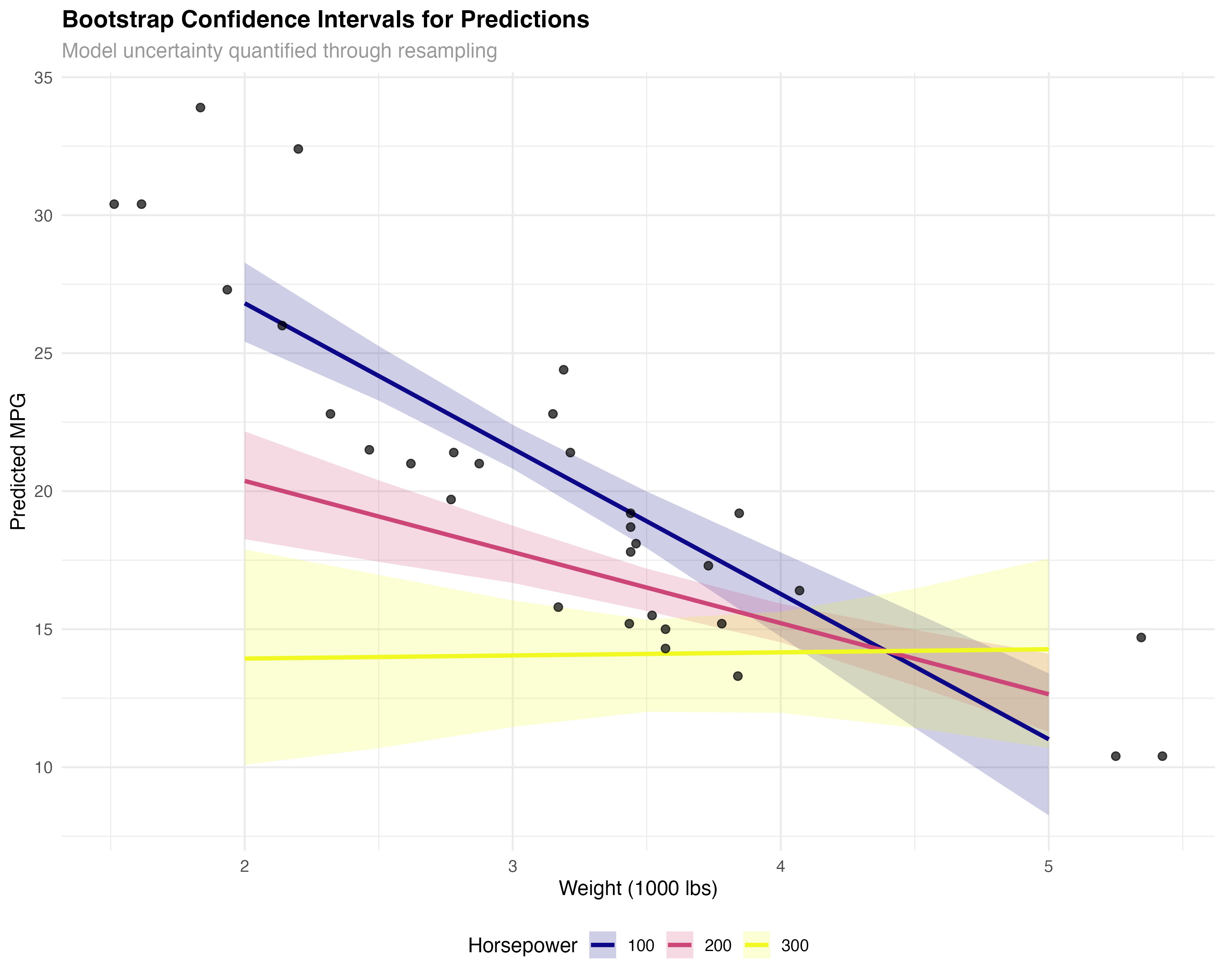
- Cross-validation: Tests predictive performance on truly unseen data
- Bootstrap: Quantifies uncertainty in parameter estimates and predictions
- Complementary: Use both for comprehensive model assessment
Model Complexity and Learning Curves
Understanding how model performance changes with complexity and sample size helps optimize both model choice and data collection strategies.
# Analyze complexity vs performance trade-off
complexity_data <- data.frame(
model = c("Simple", "Additive", "Interaction", "Complex"),
parameters = c(3, 4, 5, 6), # Including intercept and error
cv_rmse = cv_metrics$rmse,
cv_r2 = cv_metrics$r_squared,
aic = c(AIC(models$simple), AIC(models$additive),
AIC(models$interaction), AIC(models$complex))
)
ggplot(complexity_data, aes(x = parameters, y = cv_rmse)) +
geom_line(color = "#dc2626", linewidth = 1.2) +
geom_point(size = 4, color = "#2563eb") +
geom_text(aes(label = model), vjust = -1, size = 3.5, fontface = "bold") +
labs(
title = "Model Complexity vs Cross-Validation Performance",
subtitle = "Bias-variance trade-off in action",
x = "Number of Parameters",
y = "Cross-Validation RMSE"
) +
theme_minimal()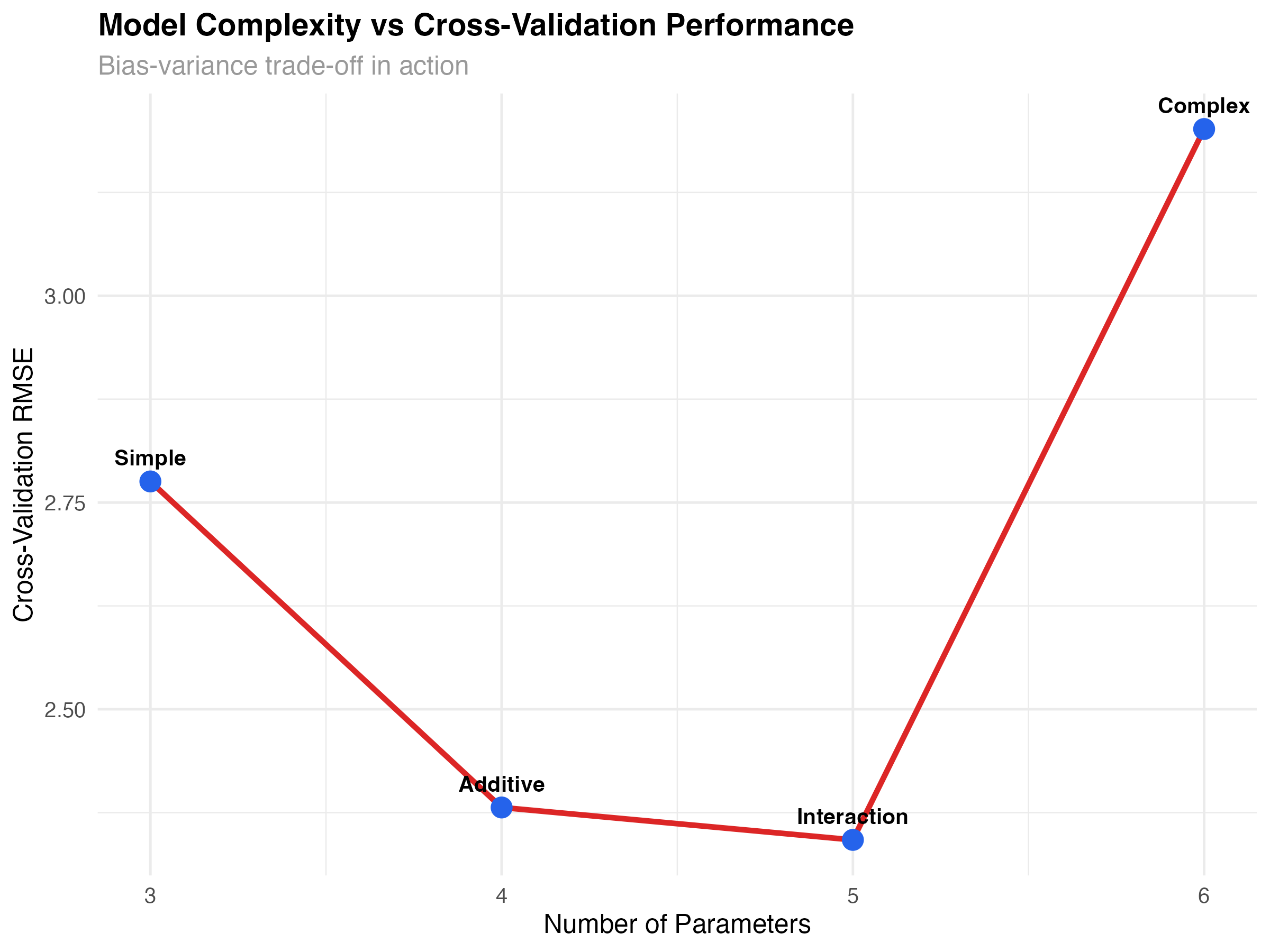
Learning Curves: Sample Size Effects
# Generate learning curve for interaction model
create_learning_curve <- function(formula, data, sizes = seq(10, nrow(data), by = 2)) {
results <- map_dfr(sizes, function(n) {
# Sample multiple times for each size
replications <- 10
metrics <- map_dfr(1:replications, function(rep) {
# Random sample of size n
sample_indices <- sample(nrow(data), n)
sample_data <- data[sample_indices, ]
# Fit model and calculate metrics
model <- glmmTMB(formula, data = sample_data)
model_data <- augment(model)
data.frame(
n = n,
rep = rep,
rmse = sqrt(mean(model_data$.resid^2)),
r_squared = 1 - var(model_data$.resid) / var(sample_data$mpg)
)
})
# Average across replications
metrics %>%
group_by(n) %>%
summarise(
rmse_mean = mean(rmse),
rmse_se = sd(rmse) / sqrt(n()),
.groups = 'drop'
)
})
}
learning_data <- create_learning_curve(mpg ~ wt * hp, mtcars)
ggplot(learning_data, aes(x = n)) +
geom_ribbon(aes(ymin = rmse_mean - rmse_se, ymax = rmse_mean + rmse_se),
alpha = 0.3, fill = "#2563eb") +
geom_line(aes(y = rmse_mean), color = "#2563eb", linewidth = 1.2) +
geom_point(aes(y = rmse_mean), color = "#2563eb", size = 2) +
labs(
title = "Learning Curve: Model Performance vs Sample Size",
subtitle = "How prediction accuracy improves with more data",
x = "Sample Size",
y = "RMSE (miles per gallon)"
) +
theme_minimal()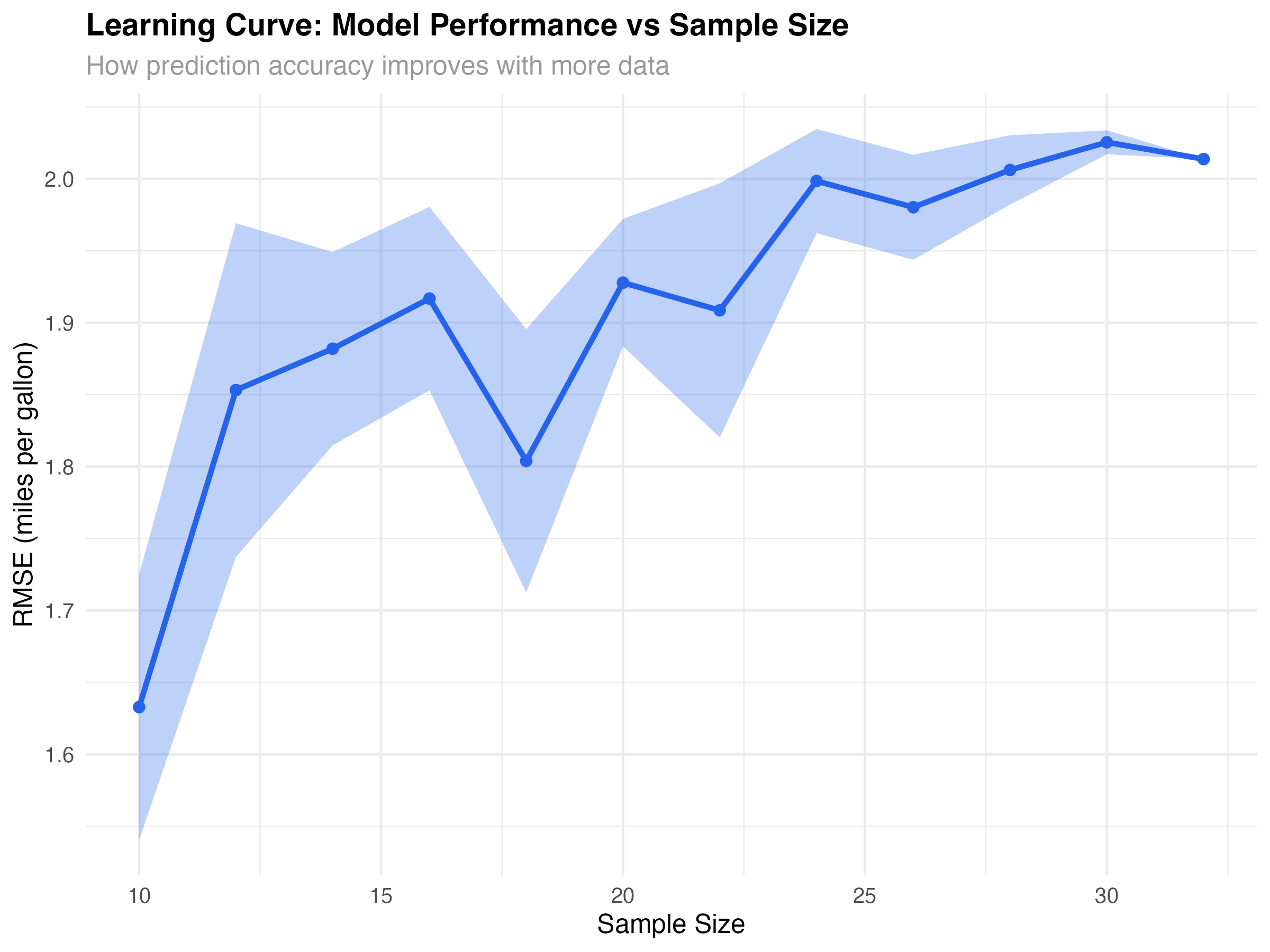
Confidence vs Prediction Intervals
Different types of uncertainty require different interval types. Understanding when to use confidence intervals vs prediction intervals is crucial for proper uncertainty communication.
# Compare confidence and prediction intervals
pred_grid_simple <- data.frame(wt = seq(2, 5, 0.1), hp = 200)
# Confidence intervals (for mean response)
conf_preds <- predict(models$interaction, newdata = pred_grid_simple, se.fit = TRUE)
pred_grid_simple$conf_fit <- conf_preds$fit
pred_grid_simple$conf_se <- conf_preds$se.fit
pred_grid_simple$conf_lower <- pred_grid_simple$conf_fit - 1.96 * pred_grid_simple$conf_se
pred_grid_simple$conf_upper <- pred_grid_simple$conf_fit + 1.96 * pred_grid_simple$conf_se
# Prediction intervals (for individual observations)
sigma <- sigma(models$interaction)
pred_grid_simple$pred_lower <- pred_grid_simple$conf_fit - 1.96 * sqrt(pred_grid_simple$conf_se^2 + sigma^2)
pred_grid_simple$pred_upper <- pred_grid_simple$conf_fit + 1.96 * sqrt(pred_grid_simple$conf_se^2 + sigma^2)
ggplot(pred_grid_simple, aes(x = wt)) +
geom_ribbon(aes(ymin = pred_lower, ymax = pred_upper),
alpha = 0.2, fill = "#dc2626") +
geom_ribbon(aes(ymin = conf_lower, ymax = conf_upper),
alpha = 0.4, fill = "#2563eb") +
geom_line(aes(y = conf_fit), color = "black", linewidth = 1.2) +
geom_point(data = filter(mtcars, hp >= 180 & hp <= 220),
aes(x = wt, y = mpg), size = 2, alpha = 0.8) +
labs(
title = "Confidence vs Prediction Intervals",
subtitle = "Blue: confidence interval for mean | Red: prediction interval for individuals",
x = "Weight (1000 lbs)",
y = "MPG",
caption = "Shown for cars with ~200 hp"
) +
theme_minimal()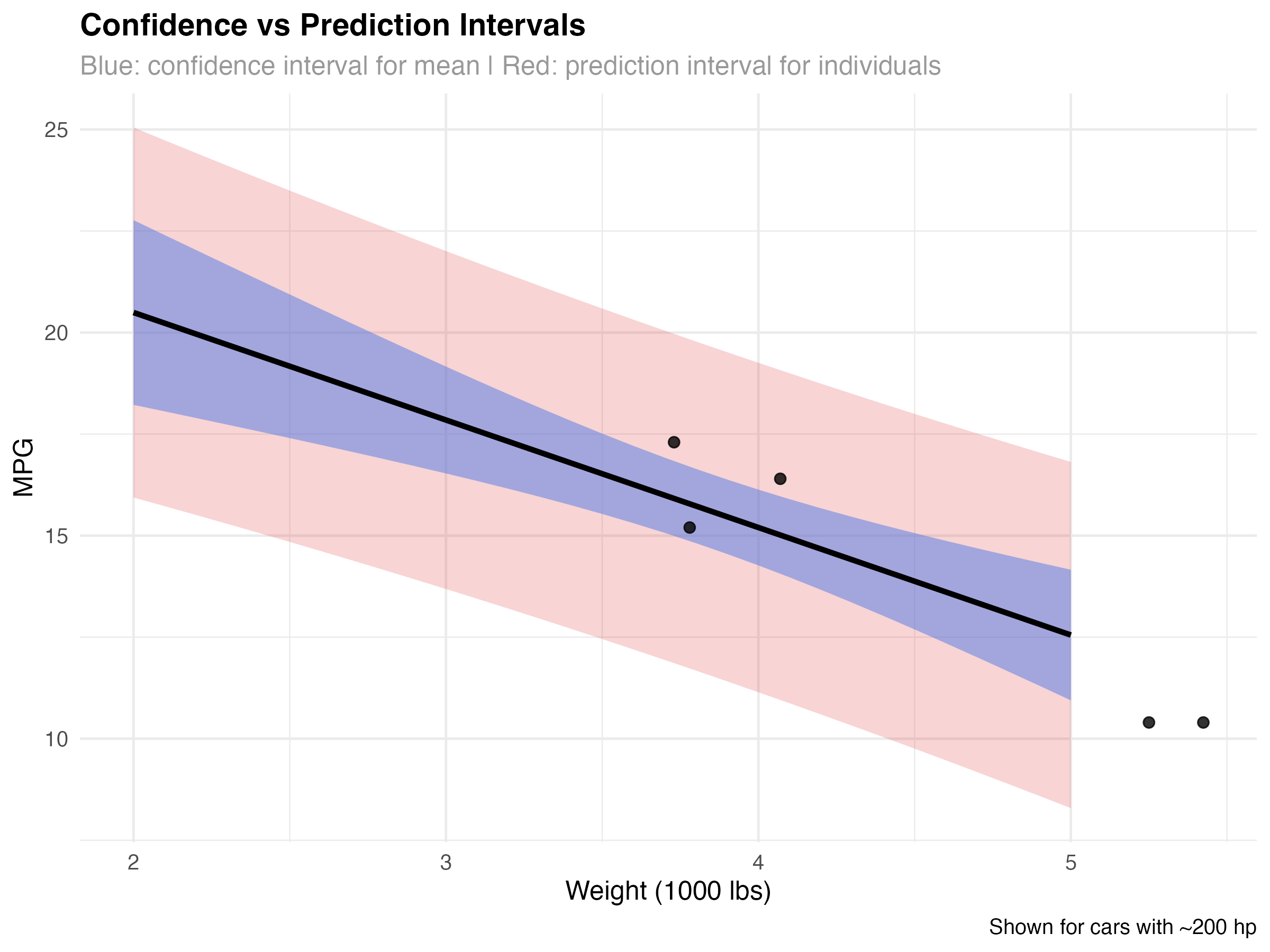
When to Use Each Interval Type
- Confidence intervals (blue): Uncertainty about the average MPG for cars with specific weight/HP combinations
- Prediction intervals (red): Expected range for an individual car's MPG
- Key difference: Prediction intervals account for both model uncertainty AND individual variation
- Practical use: Use prediction intervals when forecasting individual cases
Validation Best Practices & Summary
Complete Validation Workflow

- ✅ Best cross-validation RMSE: 2.34 MPG
- ✅ Strong prediction accuracy: R² = 0.845 on unseen data
- ✅ Optimal complexity: Best bias-variance trade-off
- ✅ Reliable uncertainty: Bootstrap intervals capture true variability
Validation Checklist for Any Model
| Validation Step | Purpose | Key Metric |
|---|---|---|
| Cross-validation | Test predictive performance | RMSE, MAE, R² |
| Predicted vs Observed plot | Visualize prediction accuracy | Points near diagonal |
| Residual analysis | Check systematic errors | Random scatter |
| Bootstrap intervals | Quantify uncertainty | Reasonable band width |
| Complexity analysis | Avoid overfitting | Optimal parameter count |
| Learning curves | Assess data sufficiency | Performance plateau |
Key Validation Principles
- Never tune on test data: Use separate validation sets or cross-validation for model selection
- Multiple metrics matter: RMSE, MAE, and R² provide different perspectives on performance
- Visualize predictions: Plots reveal patterns that summary statistics miss
- Quantify uncertainty: Provide intervals, not just point predictions
- Consider complexity costs: Simpler models often generalize better
- Validate assumptions: Combine validation with diagnostic checking
What You've Mastered
✅ Advanced Validation Skills
- Implementing cross-validation for unbiased performance assessment
- Using multiple metrics to evaluate prediction quality
- Applying bootstrap methods for uncertainty quantification
- Understanding and managing the bias-variance trade-off
- Creating learning curves to guide data collection
- Distinguishing confidence from prediction intervals
- Making evidence-based decisions about model deployment
Advanced Topics for Continued Learning
- Nested cross-validation: Proper model selection with unbiased performance estimates
- Time series validation: Forward-chaining and walk-forward analysis
- Stratified sampling: Ensuring representative validation splits
- Bayesian model averaging: Combining multiple models for better predictions
- Out-of-distribution detection: Identifying when new data differs from training
- Calibration analysis: Ensuring predicted probabilities match observed frequencies