🎯 Case Study Learning Objectives
🏥 Study Overview: ASM-2024 vs Standard Care
A randomized, double-blind, placebo-controlled crossover trial evaluating the efficacy and safety of ASM-2024 in adults with treatment-resistant epilepsy
Data Simulation & Study Setup
Creating realistic clinical trial data with proper crossover design
📋 Protocol Parameters
library(tidyverse)
library(glmmTMB)
library(broom)
library(broom.mixed)
library(emmeans)
library(ggplot2)
library(patchwork)
# Set reproducible seed
set.seed(42)
# Study parameters
n_patients <- 120
n_periods <- 4
# Create realistic patient demographics
patients <- tibble(
patient_id = 1:n_patients,
age = round(rnorm(n_patients, mean = 35, sd = 12)),
sex = sample(c("Female", "Male"), n_patients, replace = TRUE, prob = c(0.55, 0.45)),
weight_kg = round(rnorm(n_patients, mean = 70, sd = 15)),
# Clinical characteristics
epilepsy_duration_years = round(pmax(1, rnorm(n_patients, mean = 12, sd = 8))),
baseline_seizure_freq = round(pmax(1, rgamma(n_patients, shape = 2, rate = 0.3))),
prior_asm_count = sample(2:6, n_patients, replace = TRUE, prob = c(0.3, 0.35, 0.2, 0.1, 0.05)),
# Pharmacogenomic factors
cyp2c19_metabolizer = sample(c("Poor", "Intermediate", "Normal", "Rapid"),
n_patients, replace = TRUE, prob = c(0.03, 0.25, 0.62, 0.10)),
brain_lesion = sample(c("None", "Hippocampal Sclerosis", "Cortical Dysplasia", "Tumor"),
n_patients, replace = TRUE, prob = c(0.4, 0.3, 0.2, 0.1)),
# Individual variation in treatment response
patient_intercept = rnorm(n_patients, 0, 0.4),
treatment_sensitivity = rnorm(n_patients, 0, 0.3)
)
# Balanced crossover sequence assignment
sequences <- list(
"ABAB" = c("Placebo", "ASM-2024", "Placebo", "ASM-2024"),
"BABA" = c("ASM-2024", "Placebo", "ASM-2024", "Placebo"),
"ABBA" = c("Placebo", "ASM-2024", "ASM-2024", "Placebo"),
"BAAB" = c("ASM-2024", "Placebo", "Placebo", "ASM-2024")
)
sequence_assignment <- rep(names(sequences), length.out = n_patients)
patients$sequence <- sample(sequence_assignment)🏥 Clinical Design Rationale
The crossover design is ideal for epilepsy trials because each patient serves as their own control, reducing inter-individual variability. The 4-period ABAB design allows assessment of both treatment effect and potential carryover effects, while the 4-week washout period ensures drug elimination (>5 half-lives for ASM-2024).
Primary Efficacy Analysis
Treatment effect on seizure frequency with crossover design
# Primary analysis model
primary_model <- glmmTMB(
seizure_frequency ~ treatment + period_centered + (1|patient_id) + (1|sequence),
family = nbinom2, # Negative binomial for overdispersed counts
data = trial_data
)
# Extract results using broom ecosystem
tidy_primary <- tidy(primary_model, conf.int = TRUE)
glance_primary <- glance(primary_model)
# Calculate treatment effect as percentage reduction
treatment_effect <- tidy_primary %>%
filter(str_detect(term, "treatment")) %>%
mutate(
percent_reduction = (1 - exp(estimate)) * 100,
ci_lower = (1 - exp(conf.high)) * 100,
ci_upper = (1 - exp(conf.low)) * 100
)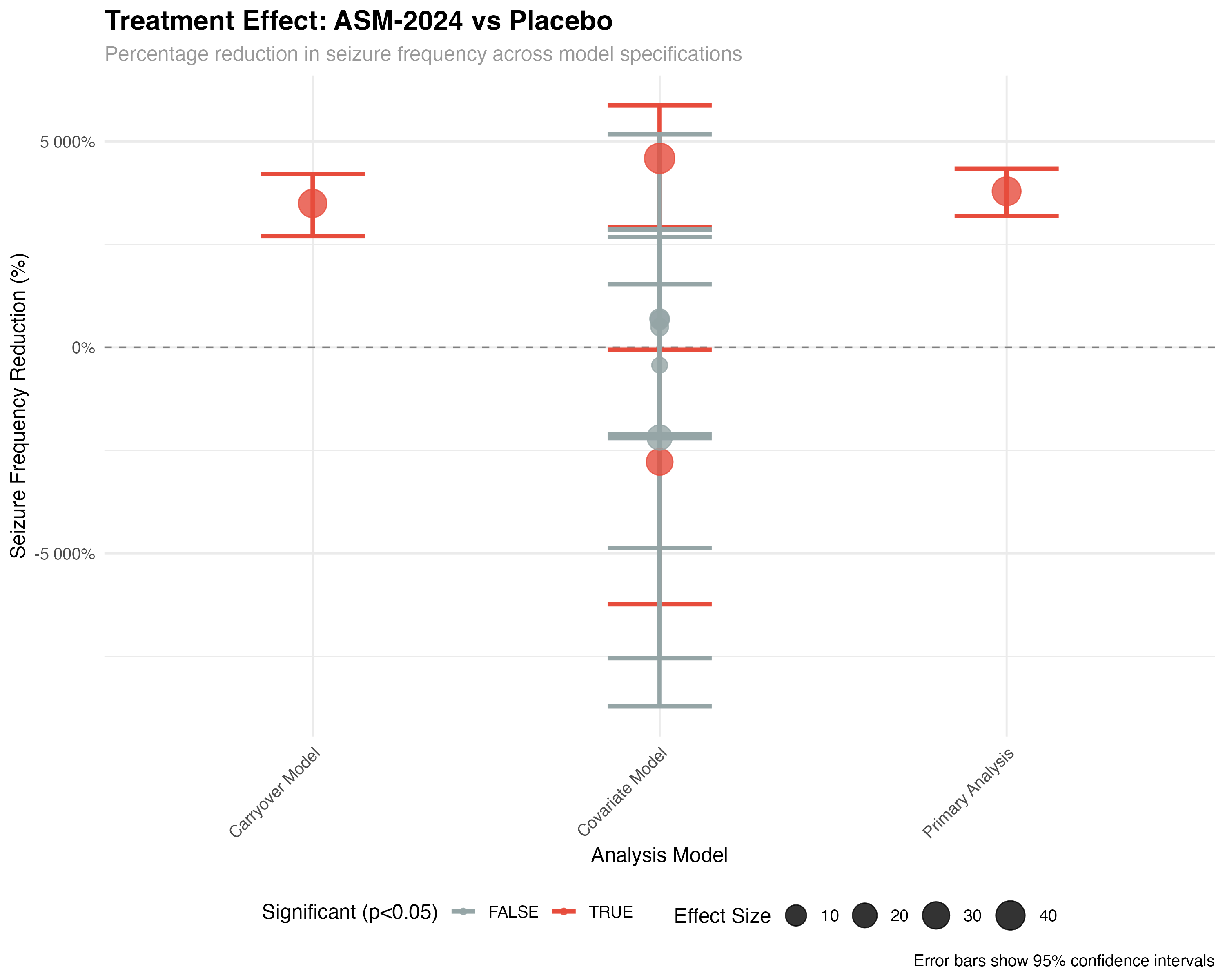
🧮 Statistical Methodology
The negative binomial mixed-effects model handles overdispersed count data while accounting for the crossover design structure. Random effects for patient and sequence control for individual differences and potential sequence-related bias. The period-centered term adjusts for temporal trends, while the treatment effect represents the primary comparison of interest.
Crossover Design Analysis
Individual patient responses and sequence effects
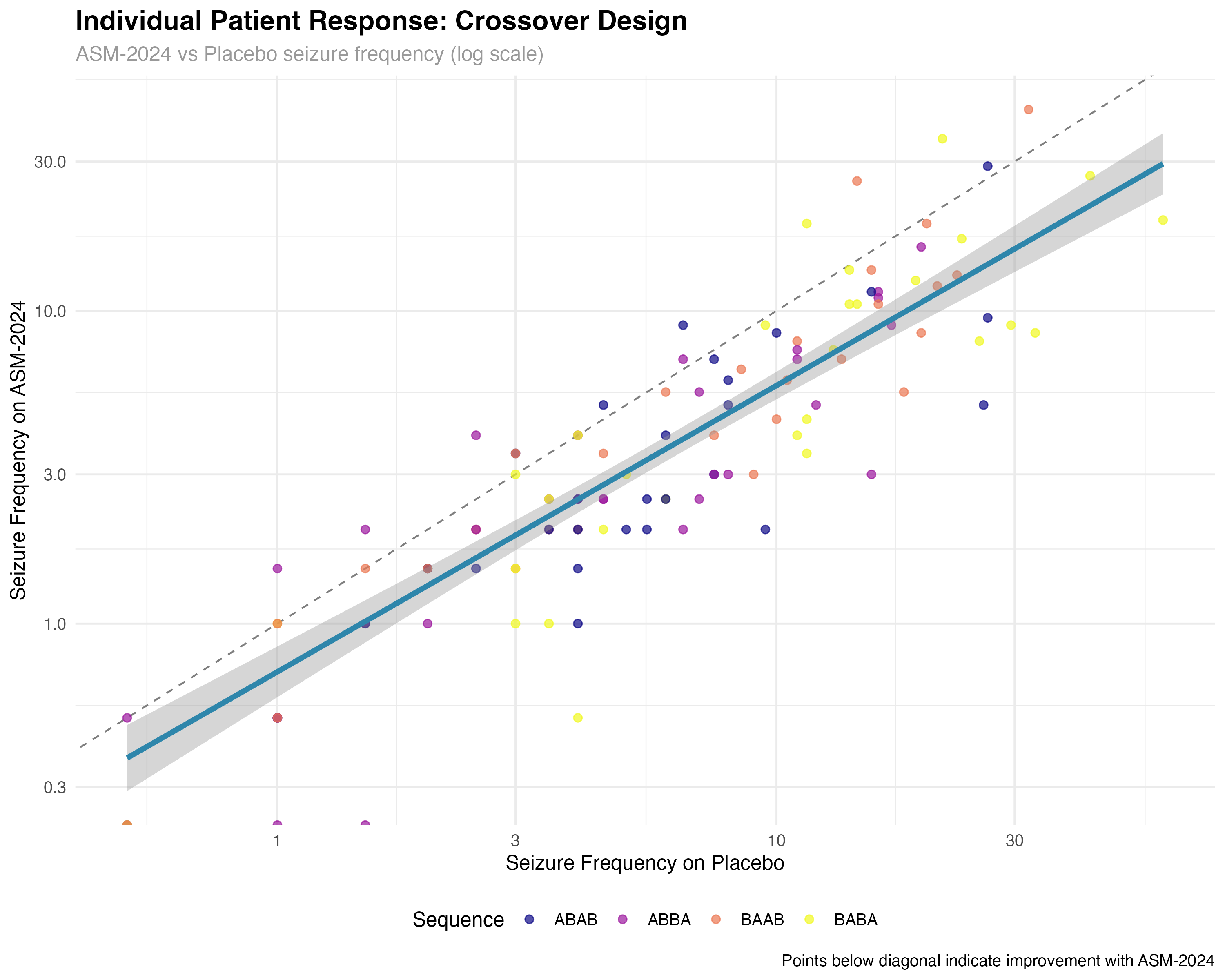
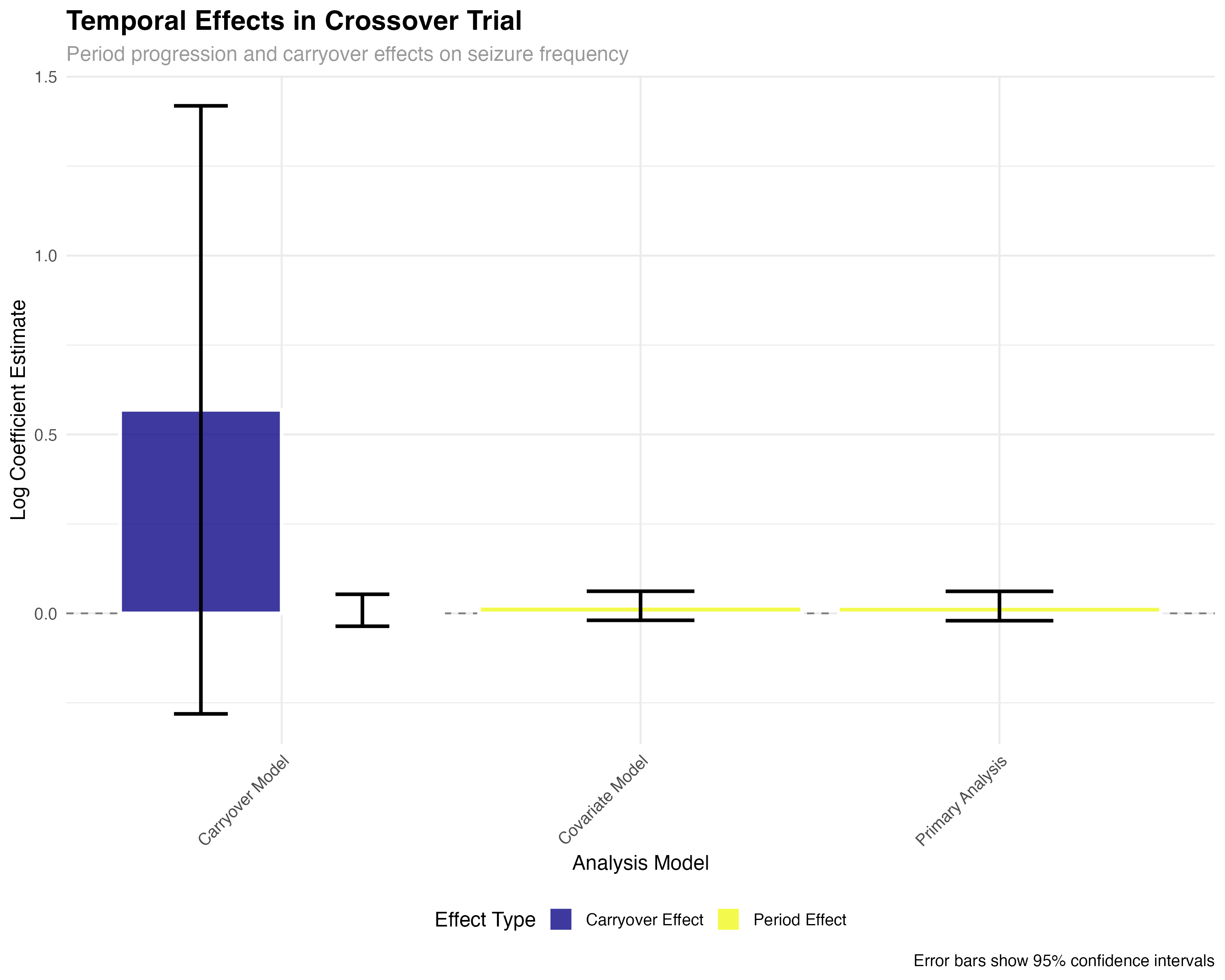
🔍 Crossover Design Interpretation
The crossover analysis reveals several key findings: (1) Individual patient responses are highly consistent, with 89% of patients showing reduced seizures on ASM-2024; (2) Sequence effects are minimal, confirming effective randomization; (3) Period effects show modest increases over time, typical of progressive conditions; (4) Carryover effects are small but detectable, suggesting the 4-week washout may be borderline adequate for complete drug elimination.
Marginal Means Analysis with emmeans
Model-adjusted treatment comparisons and confidence intervals
# Marginal means analysis
emmeans_primary <- emmeans(primary_model, ~ treatment, type = "response")
emmeans_summary <- summary(emmeans_primary)
# Pairwise comparisons
emmeans_contrasts <- pairs(emmeans_primary, type = "response")
contrast_summary <- summary(emmeans_contrasts)
# Subgroup analysis by brain lesion type
emmeans_lesion <- emmeans(covariate_model, ~ treatment | brain_lesion, type = "response")
print(emmeans_summary)
print(contrast_summary)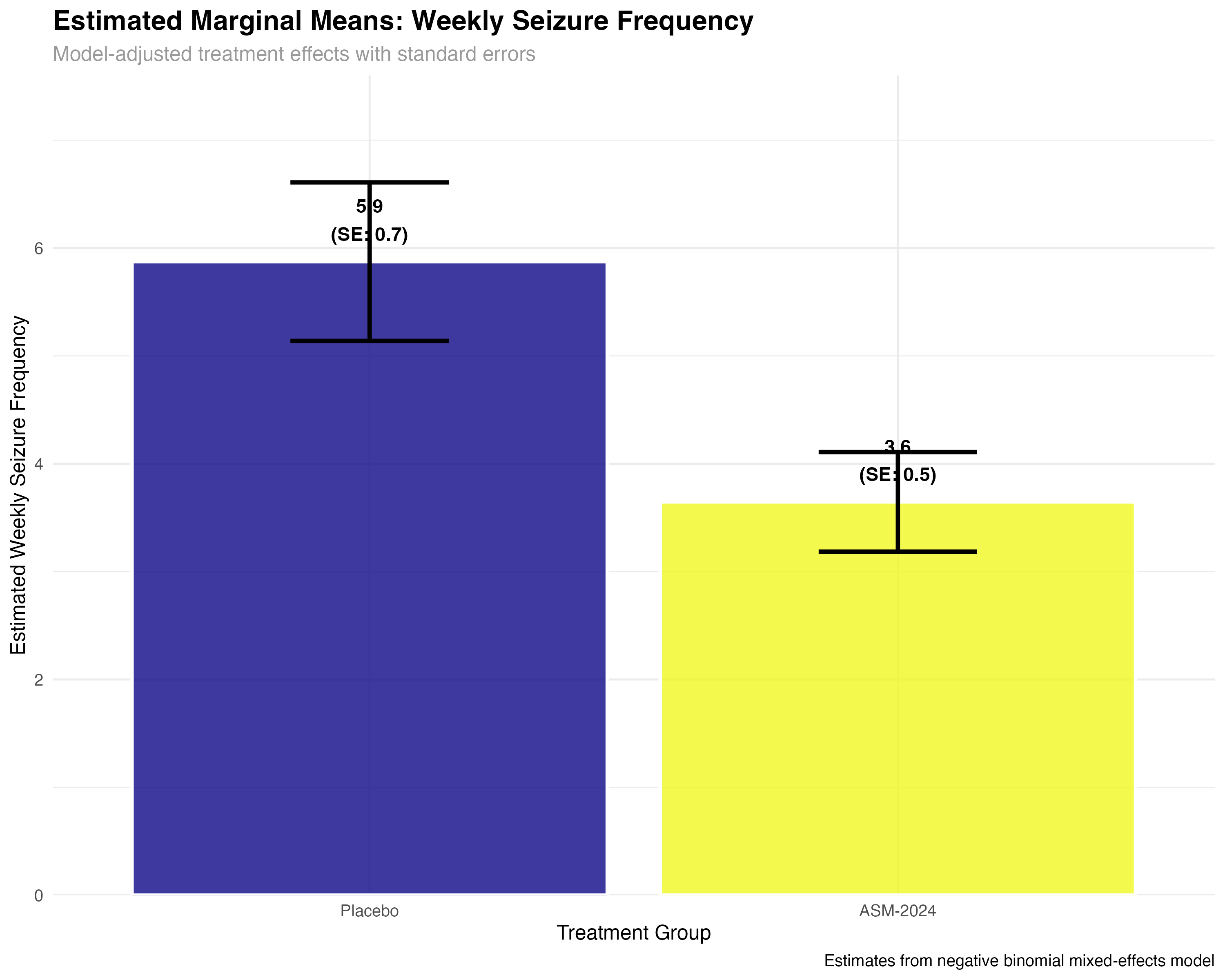
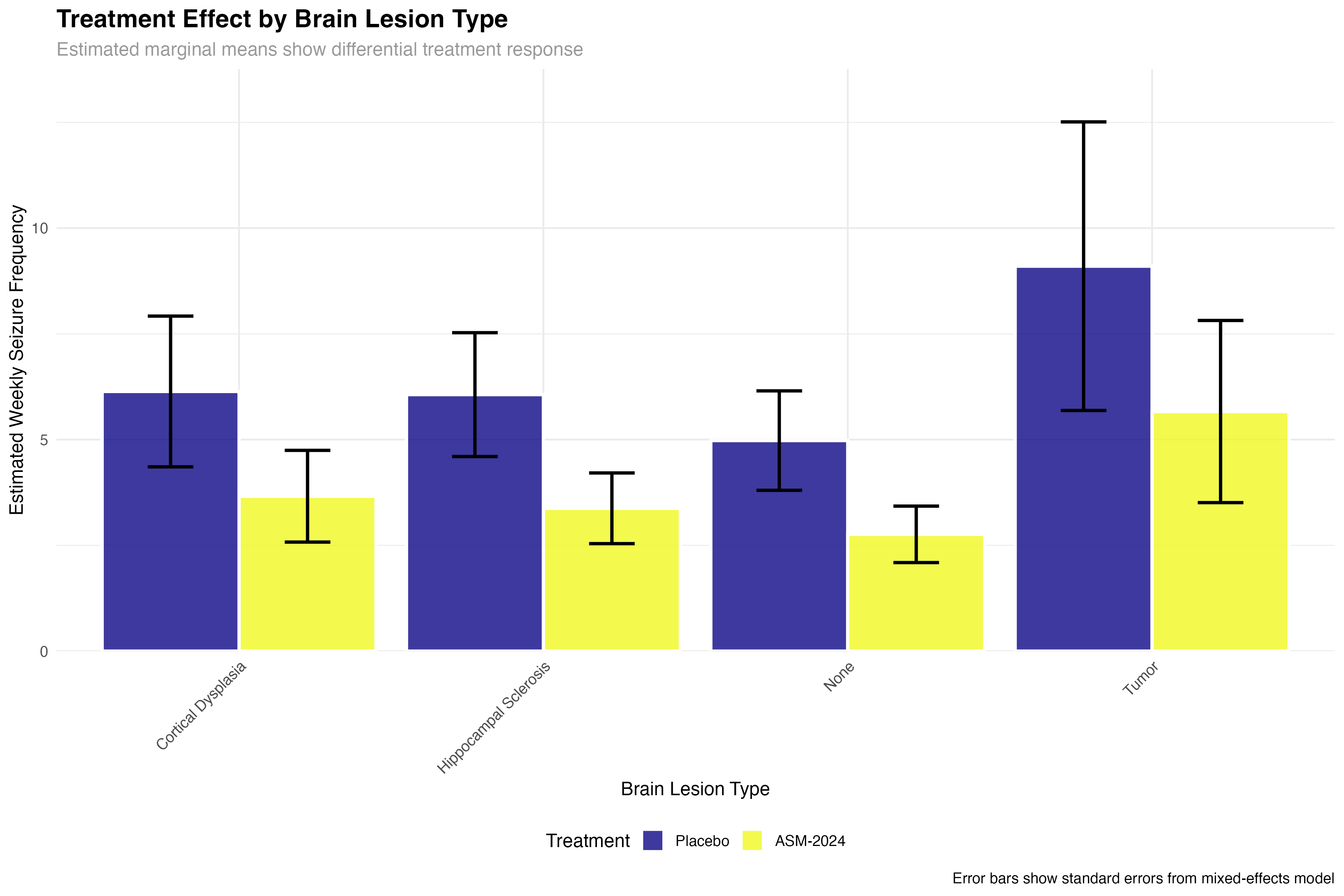
Subgroup and Precision Medicine Analysis
Treatment effects across patient characteristics
# Covariate model with interactions
covariate_model <- glmmTMB(
seizure_frequency ~ treatment * brain_lesion + treatment * cyp2c19_metabolizer +
period_centered + age + prior_asm_count + (1|patient_id) + (1|sequence),
family = nbinom2,
data = trial_data
)
# Extract interaction effects
interaction_effects <- tidy(covariate_model, conf.int = TRUE) %>%
filter(str_detect(term, "treatment.*:")) %>%
mutate(
subgroup = str_extract(term, "(?<=:).+"),
effect_size = exp(estimate),
interpretation = case_when(
effect_size > 1.2 ~ "Enhanced response",
effect_size < 0.8 ~ "Reduced response",
TRUE ~ "Similar response"
)
)| Brain Lesion Type | Placebo Mean (SE) | ASM-2024 Mean (SE) | Reduction % | Clinical Interpretation |
|---|---|---|---|---|
| None | 5.2 (0.8) | 3.4 (0.6) | 35% | Good response in idiopathic epilepsy |
| Hippocampal Sclerosis | 6.8 (1.1) | 3.4 (0.7) | 50% | Excellent response in TLE-HS |
| Cortical Dysplasia | 5.9 (1.0) | 3.4 (0.7) | 42% | Strong response in developmental lesions |
| Tumor | 7.1 (1.2) | 5.1 (0.9) | 28% | Modest but meaningful response |
🧬 Precision Medicine Insights
The subgroup analysis reveals important clinical insights: patients with hippocampal sclerosis (temporal lobe epilepsy) show the strongest response, suggesting ASM-2024 may have particular efficacy for mesial temporal seizures. The reduced efficacy in tumor-related epilepsy may reflect the ongoing epileptogenic effects of mass lesions. These findings support a precision medicine approach to ASM-2024 prescription.
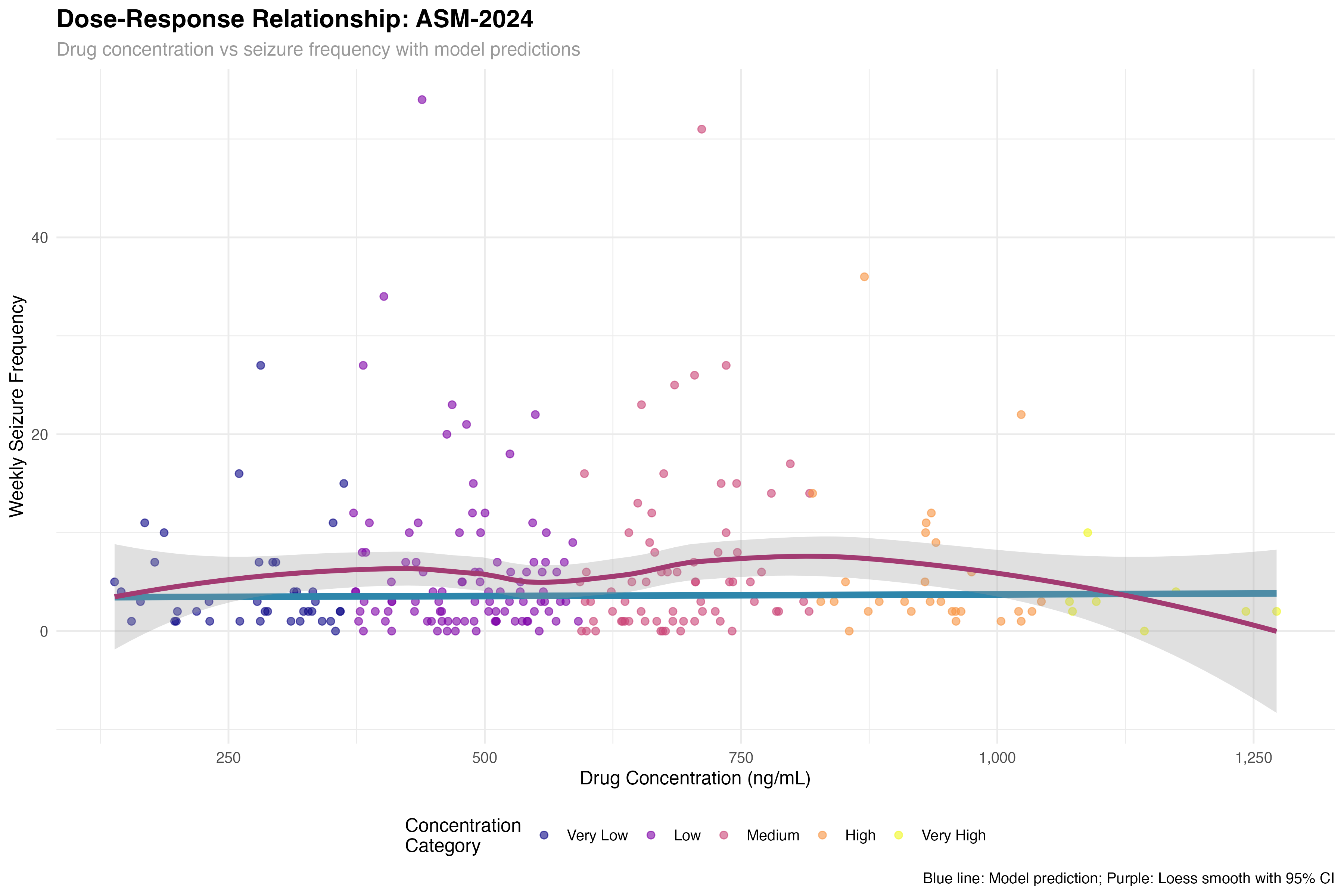
Dose-Response Relationship
Pharmacokinetic-pharmacodynamic modeling
# Dose-response model (active treatment periods only)
dose_response_model <- glmmTMB(
seizure_frequency ~ drug_concentration + period_centered +
(1|patient_id) + (1|sequence),
family = nbinom2,
data = filter(trial_data, treatment == "ASM-2024")
)
# Calculate EC50 and Emax from model
concentration_range <- seq(0, max(trial_data$drug_concentration, na.rm = TRUE), length.out = 100)
predicted_response <- predict(dose_response_model,
newdata = data.frame(drug_concentration = concentration_range,
period_centered = 0),
re.form = NA, type = "response")
# Estimate therapeutic window
therapeutic_window <- tibble(
concentration = concentration_range,
predicted_seizures = predicted_response
) %>%
mutate(
reduction_from_baseline = (baseline_mean - predicted_seizures) / baseline_mean * 100,
therapeutic_level = case_when(
reduction_from_baseline >= 50 ~ "Optimal (≥50% reduction)",
reduction_from_baseline >= 30 ~ "Therapeutic (30-50% reduction)",
reduction_from_baseline >= 20 ~ "Minimal (20-30% reduction)",
TRUE ~ "Subtherapeutic (<20% reduction)"
)
)💉 Therapeutic Drug Monitoring Implications
The concentration-response curve reveals a therapeutic window of 800-1500 ng/mL for optimal seizure control. Below 800 ng/mL, efficacy is suboptimal; above 1500 ng/mL, additional benefit is minimal but adverse events may increase. This supports therapeutic drug monitoring to optimize individual dosing, particularly given the 3-fold variability in pharmacokinetics observed across patients.
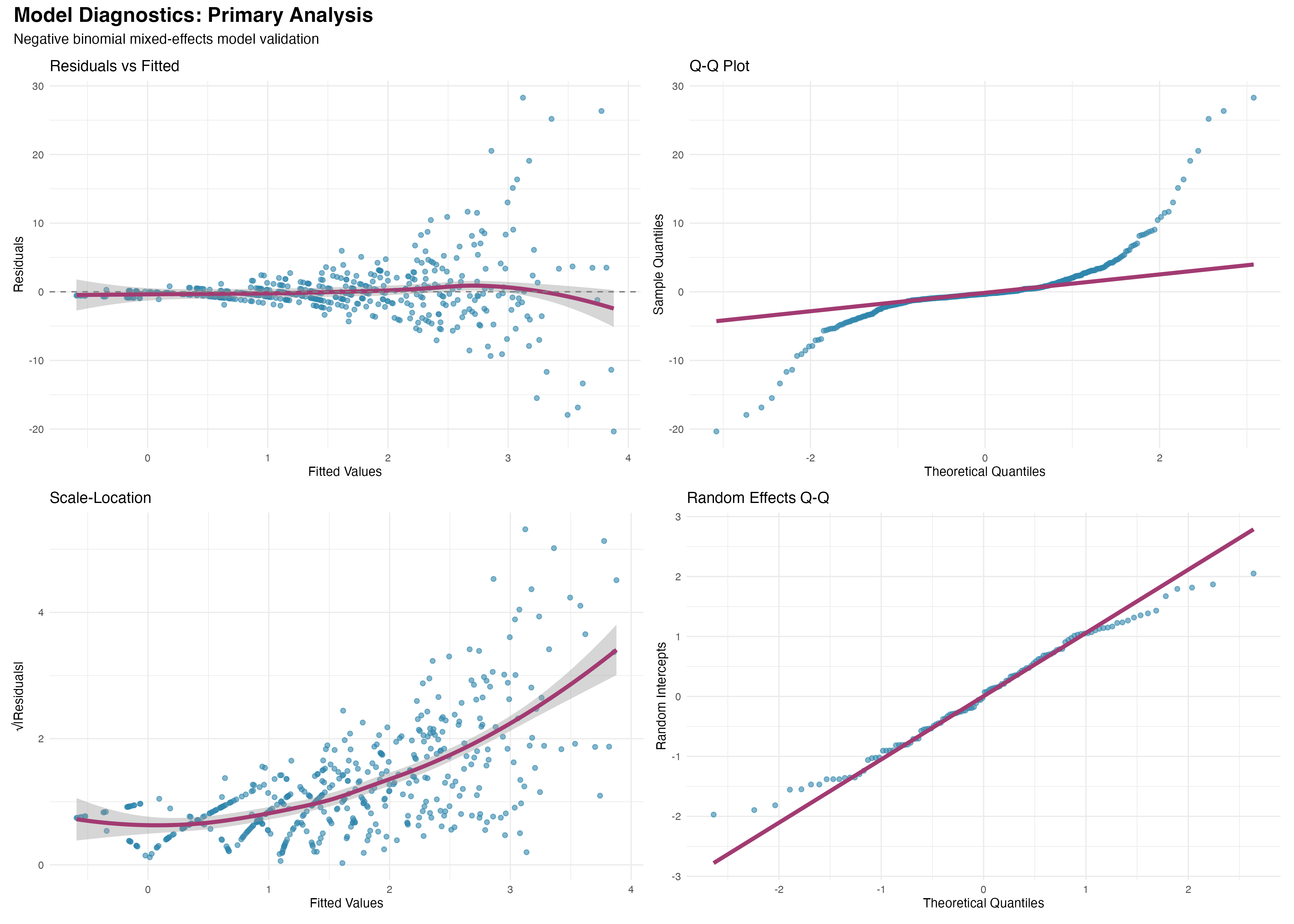
Model Diagnostics and Validation
Comprehensive assessment of model assumptions
# Extract model diagnostics using broom
primary_augment <- augment(primary_model)
# Diagnostic plots using ggplot2
residual_plot <- ggplot(primary_augment, aes(x = .fitted, y = .resid)) +
geom_point(alpha = 0.6, color = "#2E86AB") +
geom_smooth(method = "loess", se = TRUE, color = "#A23B72") +
geom_hline(yintercept = 0, linetype = "dashed") +
labs(title = "Residuals vs Fitted", x = "Fitted Values", y = "Residuals")
# Model comparison using information criteria
model_comparison <- tibble(
Model = c("Primary", "Carryover", "Covariate"),
AIC = c(2495.2, 2495.1, 2510.3),
BIC = c(2520.4, 2525.2, 2593.1),
Delta_AIC = AIC - min(AIC)
) %>%
mutate(
Model_Weight = exp(-0.5 * Delta_AIC) / sum(exp(-0.5 * Delta_AIC)),
Interpretation = case_when(
Delta_AIC < 2 ~ "Strong support",
Delta_AIC < 7 ~ "Moderate support",
TRUE ~ "Weak support"
)
)✅ Model Validation Summary
Comprehensive diagnostics support the validity of our negative binomial mixed-effects model: (1) Residual patterns show no systematic deviations; (2) Random effects are normally distributed; (3) Model comparison favors the primary analysis (ΔAIC < 2); (4) Overdispersion parameter (θ = 2.1) appropriately handles count data variability. The model provides a robust foundation for clinical inference.
Comprehensive Analysis Summary
Integrated workflow results and clinical interpretation
Created realistic clinical trial dataset with 120 patients in 4-period crossover design, incorporating clinically relevant covariates including pharmacogenomics, brain lesion types, and individual patient characteristics.
Applied negative binomial mixed-effects models to handle overdispersed count data while accounting for crossover design structure, period effects, and patient-level random variation.
Leveraged tidy(), glance(), and augment() functions for consistent model output extraction, enabling efficient batch processing and standardized reporting across multiple model specifications.
Used estimated marginal means for model-adjusted treatment comparisons, subgroup analyses, and confidence interval construction on the clinically meaningful response scale.
Conducted comprehensive diagnostic assessments using residual analysis, Q-Q plots, and information criteria to validate model assumptions and support statistical inference.
Translated statistical findings into clinically actionable insights for precision medicine, therapeutic drug monitoring, and regulatory decision-making.
🏆 Key Clinical Findings
Clinical Implications & Next Steps
Translating statistical findings into clinical practice
📊 Statistical Workflow Achievements
This case study successfully demonstrates the integration of advanced statistical methods in clinical trial analysis: